Geostatistics and kriging#
Geostatistics is the subarea of spatial statstistics which works with geostatstitical data. It finds applications in various fields such as natural resource exploration (e.g., oil and gas reserves estimation), environmental monitoring (e.g., air and water quality assessment), agriculture (e.g., soil fertility mapping), and urban planning (e.g., land use analysis) and, of course, epidemiology (e.g., disease mapping). It offers powerful tools for spatial data analysis, decision-making, and resource management in both scientific research and practical applications.
Kriging is a statistical interpolation technique used primarily in geostatistics. It is named after South African mining engineer DanielG. Krige, who developed the method in the 1950s. Kriging is employed to estimate the value of a variable at an unmeasured location based on the values observed at nearby locations.
The basic idea behind kriging is to model the spatial correlation or spatial autocorrelation of the variable being studied. This means that kriging considers the spatial structure of the data. It assumes that nearby points are more similar than those farther away and uses this information to make predictions.
Kriging provides not only the predicted value at an unmeasured location but also an estimate of the uncertainty associated with that prediction. This is valuable because it allows users to assess the reliability of the interpolated values.
As you might have guessed, kriging can be performed using Gaussian Processes! GPs are appropriate for kriging due to their flexibility in modeling complex spatial correlations, ability to quantify uncertainty and flexibility in kernel selection; i.e. GPs tick all the boxes required for kriging.
import pickle
import numpy as np
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.gaussian_process.kernels import RBF
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
import matplotlib.patches as mpatches
from matplotlib.colors import ListedColormap
import jax
import jax.numpy as jnp
import numpyro
import numpyro.distributions as dist
from numpyro.infer import init_to_median, Predictive, MCMC, NUTS
from numpyro.diagnostics import hpdi
numpyro.set_host_device_count(4) # Set the device count to enable parallel sampling
/opt/hostedtoolcache/Python/3.11.11/x64/lib/python3.11/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
Kriging: synthetic visualisation in 2d#
Let us visualise schematically how kriging looks like.
# generate synthetic data
np.random.seed(0)
x_train = np.random.uniform(0, 10, (10, 2)) # training points - x
y_train = np.sin(x_train[:, 0]) * np.cos(x_train[:, 1]) # training targets - y
plt.figure(figsize=(8, 6))
plt.scatter(x_train[:, 0], x_train[:, 1], s=150, c='none', edgecolor='red', linewidth=2, label='Training data')
plt.scatter(x_train[:, 0], x_train[:, 1], s=100, c=y_train, cmap='viridis')
plt.title('Kriging visualization in 2d')
plt.colorbar(label='y')
plt.legend()
plt.grid(True)
plt.xlim(0,10)
plt.ylim(0,10)
plt.xlabel('x1')
plt.ylabel('x2')
plt.show()
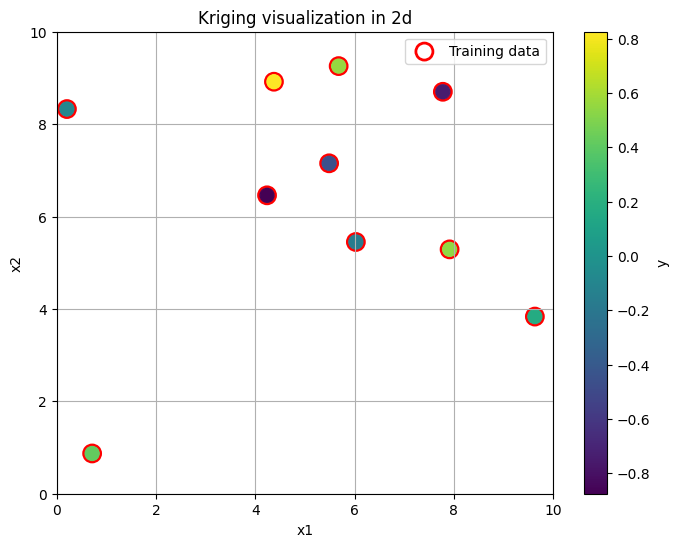
Our goal now is, given these points, to reconstruct the entire continuous surface over the region \((0,10) \times (0,10)\) assuming the GP model. Let’s do it in a non-Bayesian way first, for illustrative purpuses.
# generate grid points for visualization
x_min, x_max = 0, 10
y_min, y_max = 0, 10
xx, yy = np.meshgrid(np.linspace(x_min, x_max, 100), np.linspace(y_min, y_max, 100))
x_grid = np.c_[xx.ravel(), yy.ravel()] # 2D grid points
# fit Gaussian process model
kernel = RBF(length_scale=1.0)
gp = GaussianProcessRegressor(kernel=kernel, alpha=0.1, n_restarts_optimizer=10)
gp.fit(x_train, y_train)
# predict values for grid points
y_pred, sigma = gp.predict(x_grid, return_std=True)
plt.figure(figsize=(8, 6))
plt.scatter(xx, yy, s=100, c=y_pred.reshape(xx.shape), cmap='viridis')
plt.scatter(x_train[:, 0], x_train[:, 1], color='red', label='Training data', s=100)
plt.colorbar(label='predicted $y$')
plt.title('Kriging visualization in 2d')
plt.legend()
plt.grid(True)
plt.xlim(0,10)
plt.ylim(0,10)
plt.xlabel('x1')
plt.ylabel('x2')
plt.show()
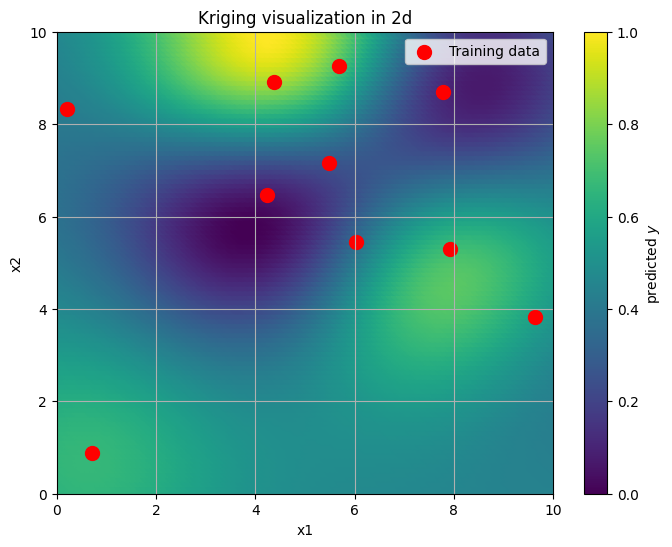
The command above GaussianProcessRegressor worked out very well. However, if we want to work with non-Gaussian likelihoods, and more complex models overall, such straightforward tools won’t be available to us. Hence, we need to understand how to implement such models in Numpyro.
1d example#
Generate data#
# synthetic data
n_points = 50
x = np.linspace(0, 2*np.pi, n_points)
f_true = np.sin(x)
# noisy observations
y_true = f_true + np.random.normal(0, 0.2, size=n_points)
# inidices to skip (and where to make predictions)
skip_idx = np.array([4, 5, 10, 15, 20, 21, 22, 25, 30, 35,36, 38, 40, 45])
# indices of observed locations excluding skip_idx
obs_idx = np.delete(np.arange(n_points), skip_idx)
y_obs = y_true[obs_idx]
Visualise#
plt.figure(figsize=(8, 6))
plt.plot(x, f_true, color='orange', label='f(x)')
plt.scatter(x[obs_idx], y_obs, color='red', label='y observed')
# unobserved locations
plt.scatter(x[skip_idx], f_true[skip_idx], color='green', label='unobserved locations', s=50, marker='D')
for idx in skip_idx:
plt.axvline(x=x[idx], color='gray', linestyle='--', linewidth=1)
plt.legend(loc='upper right')
plt.show()
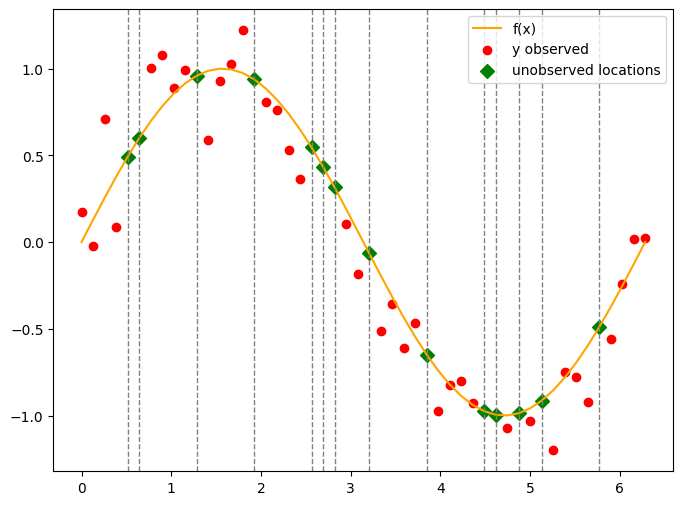
Infer#
def rbf_kernel(x1, x2, lengthscale=1.0, sigma=1.0):
sq_dist = jnp.sum(x1**2, axis=1).reshape(-1, 1) + jnp.sum(x2**2, axis=1) - 2 * jnp.dot(x1, x2.T)
K = sigma**2 * jnp.exp(-0.5 / lengthscale**2 * sq_dist)
return K
def model(x, obs_idx, y_obs=None, kernel_func=rbf_kernel, lengthcsale=0.2, jitter=1e-5):
n = x.shape[0]
K = kernel_func(x, x, lengthcsale) + jitter*jnp.eye(n)
f = numpyro.sample("f", dist.MultivariateNormal(jnp.zeros(n), covariance_matrix=K))
sigma = numpyro.sample("sigma", dist.HalfNormal(1))
numpyro.sample("y", dist.Normal(f[obs_idx], sigma), obs=y_obs)
x = jnp.linspace(0, 1, n_points).reshape(-1, 1)
nuts_kernel = NUTS(model)
mcmc = MCMC(nuts_kernel, num_samples=10000, num_warmup=2000, num_chains=2, chain_method='parallel', progress_bar=False)
mcmc.run(jax.random.PRNGKey(42), jnp.array(x), jnp.array(obs_idx), jnp.array(y_obs))
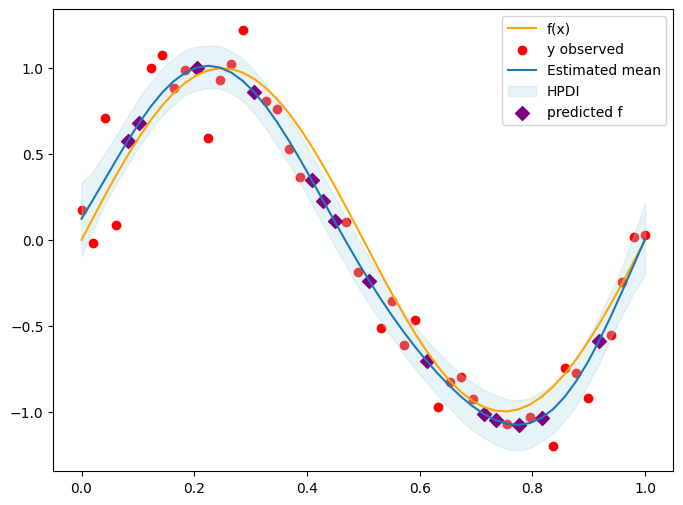
Diagnose#
mcmc.print_summary()
posterior_samples = mcmc.get_samples()
f_posterior = posterior_samples['f']
f_mean = jnp.mean(f_posterior, axis=0)
f_hpdi = hpdi(f_posterior, 0.90)
plt.figure(figsize=(8, 6))
plt.plot(x, f_true, color='orange', label='f(x)')
plt.scatter(x[obs_idx], y_obs, color='red', label='y observed')
plt.plot(x, f_mean, label='Estimated mean')
plt.fill_between(x.squeeze(), f_hpdi[0], f_hpdi[1], color='lightblue', alpha=0.3, label='HPDI') # Uncertainty bounds
plt.scatter(x[skip_idx], f_mean[skip_idx], color='purple', label='predicted f', s=50, marker='D')
plt.legend()
plt.show()
mean std median 5.0% 95.0% n_eff r_hat
f[0] 0.12 0.13 0.12 -0.09 0.33 4225.91 1.00
f[1] 0.23 0.10 0.23 0.06 0.40 3792.92 1.00
f[2] 0.35 0.09 0.35 0.20 0.50 3382.63 1.00
f[3] 0.46 0.09 0.46 0.33 0.61 3034.96 1.00
f[4] 0.57 0.09 0.58 0.44 0.72 2805.64 1.00
f[5] 0.68 0.09 0.68 0.54 0.83 2680.45 1.00
f[6] 0.77 0.09 0.78 0.64 0.92 2607.97 1.00
f[7] 0.86 0.08 0.86 0.72 1.00 2520.06 1.00
f[8] 0.92 0.08 0.92 0.79 1.06 2451.53 1.00
f[9] 0.97 0.08 0.97 0.84 1.10 2413.03 1.00
f[10] 1.00 0.08 1.00 0.87 1.13 2421.14 1.00
f[11] 1.01 0.08 1.01 0.88 1.13 2488.65 1.00
f[12] 1.00 0.07 1.00 0.88 1.13 2603.09 1.00
f[13] 0.98 0.07 0.98 0.86 1.10 2635.56 1.00
f[14] 0.93 0.08 0.93 0.81 1.06 2631.18 1.00
f[15] 0.86 0.08 0.86 0.74 0.99 2583.48 1.00
f[16] 0.78 0.08 0.78 0.65 0.91 2508.25 1.00
f[17] 0.69 0.08 0.69 0.55 0.81 2416.01 1.00
f[18] 0.58 0.08 0.58 0.45 0.72 2335.34 1.00
f[19] 0.47 0.08 0.47 0.33 0.61 2279.03 1.00
f[20] 0.35 0.09 0.35 0.21 0.49 2249.53 1.00
f[21] 0.23 0.09 0.23 0.09 0.37 2250.98 1.00
f[22] 0.11 0.09 0.11 -0.04 0.25 2284.85 1.00
f[23] -0.01 0.09 -0.01 -0.15 0.13 2330.71 1.00
f[24] -0.13 0.08 -0.13 -0.27 0.01 2419.46 1.00
f[25] -0.24 0.08 -0.24 -0.38 -0.10 2521.25 1.00
f[26] -0.34 0.08 -0.34 -0.47 -0.21 2654.44 1.00
f[27] -0.44 0.08 -0.44 -0.57 -0.31 2844.05 1.00
f[28] -0.54 0.08 -0.54 -0.67 -0.42 3067.37 1.00
f[29] -0.62 0.08 -0.62 -0.75 -0.51 3088.45 1.00
f[30] -0.71 0.08 -0.70 -0.83 -0.59 3036.02 1.00
f[31] -0.78 0.08 -0.78 -0.91 -0.65 2838.69 1.00
f[32] -0.85 0.08 -0.85 -0.98 -0.72 2610.11 1.00
f[33] -0.91 0.08 -0.91 -1.05 -0.79 2433.56 1.00
f[34] -0.97 0.08 -0.97 -1.11 -0.83 2337.99 1.00
f[35] -1.02 0.09 -1.01 -1.16 -0.87 2300.48 1.00
f[36] -1.05 0.09 -1.05 -1.19 -0.90 2307.49 1.00
f[37] -1.07 0.09 -1.07 -1.22 -0.93 2359.16 1.00
f[38] -1.08 0.09 -1.08 -1.23 -0.93 2441.15 1.00
f[39] -1.07 0.09 -1.07 -1.21 -0.92 2544.63 1.00
f[40] -1.04 0.09 -1.04 -1.17 -0.88 2642.85 1.00
f[41] -0.99 0.09 -0.99 -1.13 -0.84 2670.14 1.00
f[42] -0.91 0.09 -0.91 -1.05 -0.77 2705.11 1.00
f[43] -0.82 0.09 -0.82 -0.97 -0.69 2697.70 1.00
f[44] -0.71 0.08 -0.72 -0.85 -0.57 2692.75 1.00
f[45] -0.59 0.08 -0.59 -0.72 -0.45 2756.15 1.00
f[46] -0.45 0.08 -0.45 -0.57 -0.31 2935.85 1.00
f[47] -0.30 0.09 -0.30 -0.44 -0.15 3207.06 1.00
f[48] -0.15 0.10 -0.15 -0.31 0.02 3595.31 1.00
f[49] 0.01 0.13 0.01 -0.20 0.22 3968.51 1.00
sigma 0.20 0.03 0.19 0.15 0.24 3669.82 1.00
Number of divergences: 0
xmin = np.min([np.min(f_true), np.min(f_mean)])
xmax = np.max([np.max(f_true), np.max(f_mean)])
ymin = xmin
ymax = xmax
plt.figure(figsize=(4, 4))
plt.plot(f_true[skip_idx], f_mean[skip_idx], 'o', label='Obtained results')
plt.plot([xmin, xmax], [ymin, ymax], color='red', linestyle='--', label='Ideal prediction')
plt.xlabel('True $f$ at unobserved locations')
plt.ylabel('Predicted $f$ at unobserved locations')
plt.xlim(xmin, xmax)
plt.ylim(ymin, ymax)
plt.title('Comparison of true and predicted $f$')
plt.legend()
plt.grid(True)
plt.show()
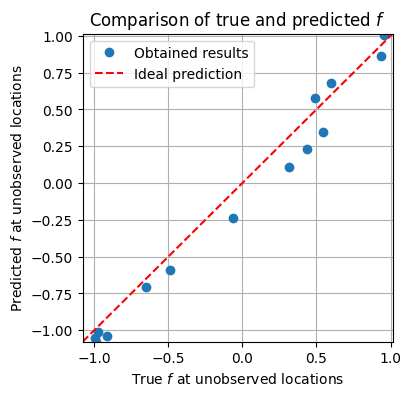
2d example#
Generate data#
n_points_x = 10
n_points_y = 8
x = jnp.linspace(0, 2*jnp.pi, n_points_x)
y = jnp.linspace(0, 2*jnp.pi, n_points_y)
xx, yy = jnp.meshgrid(x, y)
x_2d = jnp.column_stack([xx.ravel(), yy.ravel()])
skip_idx = [(0, 1), (2, 4), (3, 1), (5,6)]
obs_idx = np.delete(np.arange(n_points_x * n_points_y), [i * n_points_x + j for i, j in skip_idx])
f_true = jnp.sin(xx/1.2) * jnp.cos(yy)
noise = np.random.normal(0, 0.1, size=(n_points_y, n_points_x))
y_true = (f_true + noise)
Visualise#
cmap = plt.cm.viridis
cmap.set_bad(color='red')
fig, axes = plt.subplots(1, 2, figsize=(10, 6))
im1 = axes[0].imshow(f_true, extent=(0, 2*np.pi, 0, 2*np.pi), origin='lower', cmap='viridis')
axes[0].set_title('True Function $f(\cdot)$')
axes[0].set_xlabel('x')
axes[0].set_ylabel('y')
masked_data = np.ma.masked_where(np.zeros_like(y_true, dtype=bool), y_true)
for idx in skip_idx:
masked_data[idx] = np.ma.masked
im2 = axes[1].imshow(masked_data, extent=(0, 2*np.pi, 0, 2*np.pi), origin='lower', cmap=cmap)
axes[1].set_title('Observed Data')
axes[1].set_xlabel('x')
axes[1].set_ylabel('y')
legend_handles = [mpatches.Patch(color='red', label='Missing Values')]
axes[1].legend(handles=legend_handles)
cax = fig.add_axes([1.02, 0.15, 0.02, 0.7]) # [left, bottom, width, height]
cbar = fig.colorbar(im1, cax=cax)
cbar.set_label('Value')
plt.tight_layout()
plt.show()
/tmp/ipykernel_3019/1594027620.py:27: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
plt.tight_layout()
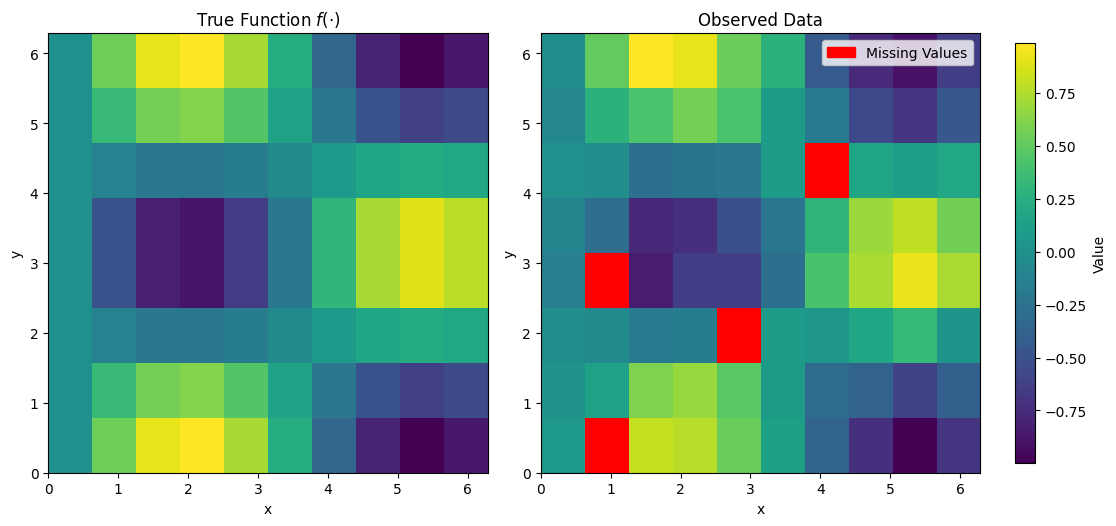
Infer#
y_true_flat = y_true.ravel()
y_obs = y_true_flat[obs_idx]
print(y_true.shape)
print(y_true.shape)
# check the shapes
print(x_2d.shape)
print(obs_idx.shape)
print(y_obs.shape)
(8, 10)
(8, 10)
(80, 2)
(76,)
(76,)
# ATTENTION: this cell might take a while to run
nuts_kernel = NUTS(model)
mcmc = MCMC(nuts_kernel, num_samples=10000, num_warmup=2000, num_chains=2, chain_method='parallel', progress_bar=False)
mcmc.run(jax.random.PRNGKey(42), jnp.array(x_2d), jnp.array(obs_idx), jnp.array(y_obs))
# Print summary statistics of posterior
mcmc.print_summary()
# Get the posterior samples
posterior_samples = mcmc.get_samples()
f_posterior = posterior_samples['f']
# Calculate mean and standard deviation
f_mean = jnp.mean(f_posterior, axis=0)
f_std = jnp.std(f_posterior, axis=0)
mean std median 5.0% 95.0% n_eff r_hat
f[0] 0.11 0.15 0.11 -0.11 0.35 40701.69 1.00
f[1] 0.00 1.01 0.01 -1.66 1.61 7779.13 1.00
f[2] 0.94 0.15 0.95 0.70 1.17 18361.93 1.00
f[3] 0.90 0.15 0.91 0.67 1.14 22819.84 1.00
f[4] 0.63 0.14 0.63 0.39 0.86 24751.55 1.00
f[5] 0.17 0.15 0.17 -0.08 0.40 35942.61 1.00
f[6] -0.36 0.14 -0.37 -0.60 -0.14 36100.24 1.00
f[7] -0.78 0.15 -0.79 -1.01 -0.54 23083.05 1.00
f[8] -1.09 0.15 -1.10 -1.32 -0.85 7730.31 1.00
f[9] -0.76 0.15 -0.77 -0.99 -0.51 25041.35 1.00
f[10] 0.04 0.15 0.05 -0.20 0.28 40492.65 1.00
f[11] 0.19 0.14 0.19 -0.04 0.42 33194.49 1.00
f[12] 0.71 0.15 0.71 0.47 0.93 28907.70 1.00
f[13] 0.79 0.15 0.80 0.54 1.01 27949.14 1.00
f[14] 0.56 0.15 0.56 0.32 0.79 27966.18 1.00
f[15] 0.12 0.15 0.12 -0.13 0.35 32633.67 1.00
f[16] -0.31 0.15 -0.32 -0.54 -0.07 32397.39 1.00
f[17] -0.39 0.15 -0.39 -0.61 -0.13 30275.34 1.00
f[18] -0.65 0.15 -0.66 -0.89 -0.42 25864.74 1.00
f[19] -0.41 0.15 -0.41 -0.65 -0.18 37767.32 1.00
f[20] 0.02 0.15 0.02 -0.21 0.26 37786.58 1.00
f[21] -0.03 0.14 -0.02 -0.26 0.20 37034.29 1.00
f[22] -0.17 0.15 -0.17 -0.40 0.08 39593.65 1.00
f[23] -0.15 0.15 -0.15 -0.38 0.09 36813.04 1.00
f[24] 0.01 1.00 0.01 -1.68 1.63 4853.25 1.00
f[25] 0.12 0.15 0.13 -0.11 0.38 37573.57 1.00
f[26] 0.09 0.15 0.09 -0.14 0.33 33937.56 1.00
f[27] 0.21 0.15 0.21 -0.02 0.45 37656.77 1.00
f[28] 0.40 0.15 0.41 0.15 0.63 34908.67 1.00
f[29] 0.06 0.15 0.06 -0.17 0.29 30314.75 1.00
f[30] -0.12 0.14 -0.13 -0.35 0.11 31233.06 1.00
f[31] -0.01 1.00 -0.02 -1.64 1.64 6640.97 1.00
f[32] -0.93 0.15 -0.94 -1.15 -0.68 16530.09 1.00
f[33] -0.68 0.15 -0.69 -0.90 -0.43 28224.95 1.00
f[34] -0.68 0.15 -0.69 -0.91 -0.44 24443.35 1.00
f[35] -0.28 0.15 -0.28 -0.52 -0.04 33157.94 1.00
f[36] 0.49 0.15 0.50 0.26 0.73 29425.54 1.00
f[37] 0.85 0.15 0.86 0.62 1.09 20069.92 1.00
f[38] 1.06 0.15 1.08 0.83 1.30 10927.85 1.00
f[39] 0.85 0.15 0.86 0.61 1.08 18351.04 1.00
f[40] -0.08 0.14 -0.09 -0.31 0.15 37058.94 1.00
f[41] -0.30 0.15 -0.30 -0.53 -0.06 32162.22 1.00
f[42] -0.83 0.15 -0.84 -1.08 -0.60 12467.18 1.00
f[43] -0.79 0.15 -0.80 -1.02 -0.55 16624.65 1.00
f[44] -0.55 0.14 -0.56 -0.78 -0.31 26848.25 1.00
f[45] -0.22 0.14 -0.22 -0.46 0.01 29257.08 1.00
f[46] 0.36 0.15 0.37 0.13 0.60 33842.84 1.00
f[47] 0.80 0.15 0.81 0.55 1.02 26162.56 1.00
f[48] 0.92 0.15 0.93 0.67 1.15 20842.35 1.00
f[49] 0.66 0.14 0.66 0.42 0.88 25902.65 1.00
f[50] 0.03 0.15 0.03 -0.20 0.26 35047.76 1.00
f[51] 0.01 0.14 0.01 -0.23 0.24 36325.74 1.00
f[52] -0.27 0.14 -0.27 -0.50 -0.03 36610.49 1.00
f[53] -0.23 0.15 -0.23 -0.46 0.00 28270.52 1.00
f[54] -0.20 0.15 -0.20 -0.44 0.03 37544.16 1.00
f[55] 0.13 0.15 0.13 -0.11 0.37 37456.54 1.00
f[56] -0.02 1.00 -0.03 -1.71 1.56 8495.16 1.00
f[57] 0.21 0.14 0.22 -0.02 0.45 40347.86 1.00
f[58] 0.14 0.15 0.14 -0.11 0.37 31973.34 1.00
f[59] 0.24 0.15 0.24 -0.01 0.48 31664.17 1.00
f[60] -0.07 0.15 -0.07 -0.31 0.16 36232.42 1.00
f[61] 0.34 0.15 0.34 0.09 0.57 31435.44 1.00
f[62] 0.50 0.15 0.50 0.26 0.74 36586.30 1.00
f[63] 0.67 0.15 0.67 0.43 0.90 28928.81 1.00
f[64] 0.50 0.14 0.51 0.27 0.73 32731.22 1.00
f[65] 0.12 0.15 0.12 -0.11 0.36 29691.02 1.00
f[66] -0.17 0.15 -0.17 -0.40 0.07 33482.95 1.00
f[67] -0.60 0.14 -0.60 -0.82 -0.36 24153.00 1.00
f[68] -0.76 0.15 -0.76 -0.99 -0.52 23800.98 1.00
f[69] -0.49 0.14 -0.49 -0.72 -0.25 31553.91 1.00
f[70] 0.01 0.15 0.02 -0.22 0.25 35218.40 1.00
f[71] 0.60 0.15 0.61 0.35 0.83 24580.46 1.00
f[72] 1.13 0.15 1.15 0.89 1.36 12208.61 1.00
f[73] 1.06 0.15 1.07 0.80 1.29 11201.84 1.00
f[74] 0.62 0.14 0.63 0.39 0.86 31291.79 1.00
f[75] 0.33 0.15 0.34 0.11 0.58 30885.87 1.00
f[76] -0.46 0.15 -0.47 -0.70 -0.22 30952.18 1.00
f[77] -0.83 0.15 -0.84 -1.08 -0.61 18131.48 1.00
f[78] -0.98 0.15 -1.00 -1.22 -0.74 13211.91 1.00
f[79] -0.68 0.15 -0.69 -0.91 -0.44 30689.59 1.00
sigma 0.13 0.08 0.11 0.02 0.24 198.76 1.01
Number of divergences: 0
f_mean_2d = f_mean.reshape(xx.shape)
f_std_2d = f_std.reshape(xx.shape)
plt.figure(figsize=(20, 6))
plt.subplot(1, 3, 1)
plt.imshow(f_true, extent=(0, 2*np.pi, 0, 2*np.pi), origin='lower', cmap='viridis')
plt.colorbar()
plt.title('True Function')
plt.subplot(1, 3, 2)
plt.imshow(f_mean_2d, extent=(0, 2*np.pi, 0, 2*np.pi), origin='lower', cmap='viridis')
plt.colorbar()
plt.title('Mean Prediction')
plt.subplot(1, 3, 3)
plt.imshow(f_std_2d, extent=(0, 2*np.pi, 0, 2*np.pi), origin='lower', cmap='viridis')
plt.colorbar()
plt.title('Std')
plt.suptitle('Kriging 2d with Numpyro')
plt.show()
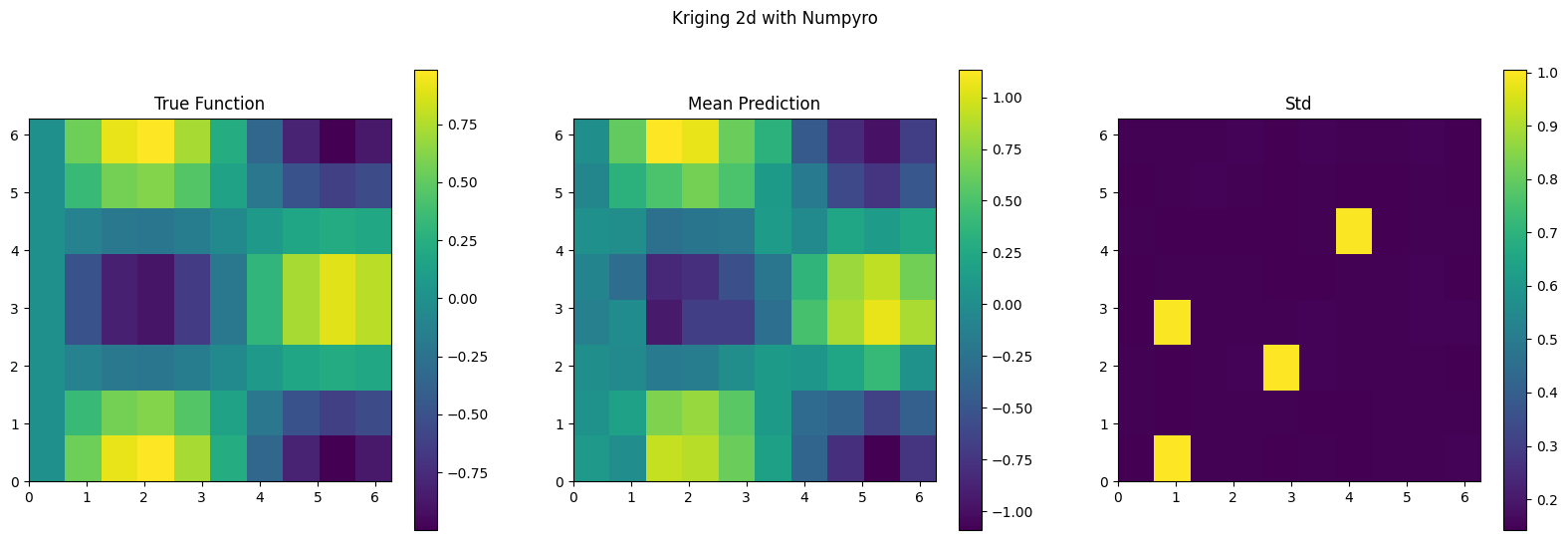
f_true_flat = f_true.flatten()
xmin = np.min([np.min(f_true_flat), np.min(f_mean)])
xmax = np.max([np.max(f_true_flat), np.max(f_mean)])
ymin = xmin
ymax = xmax
plt.figure(figsize=(4, 4))
skip_idx_1d = np.array([i * n_points_x + j for i, j in skip_idx])
plt.plot(f_true_flat[skip_idx_1d], f_mean[skip_idx_1d], 'o', label='Obtained results')
plt.plot([xmin, xmax], [ymin, ymax], color='red', linestyle='--', label='Ideal prediction')
plt.xlabel('True $f$ at unobserved locations')
plt.ylabel('Predicted $f$ at unobserved locations')
plt.xlim(xmin, xmax)
plt.ylim(ymin, ymax)
plt.title('Comparison of True and Predicted $f$')
plt.legend()
plt.grid(True)
plt.show()
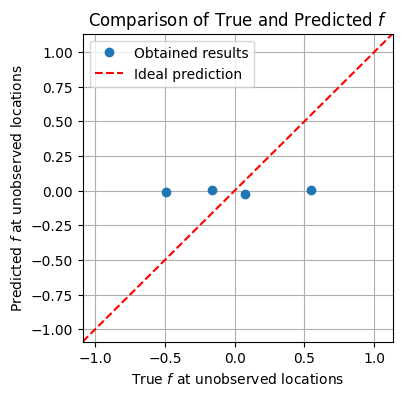
Group Task
It does not look like the model has done a good job estimating unobserved values.
What could have gone wrong?
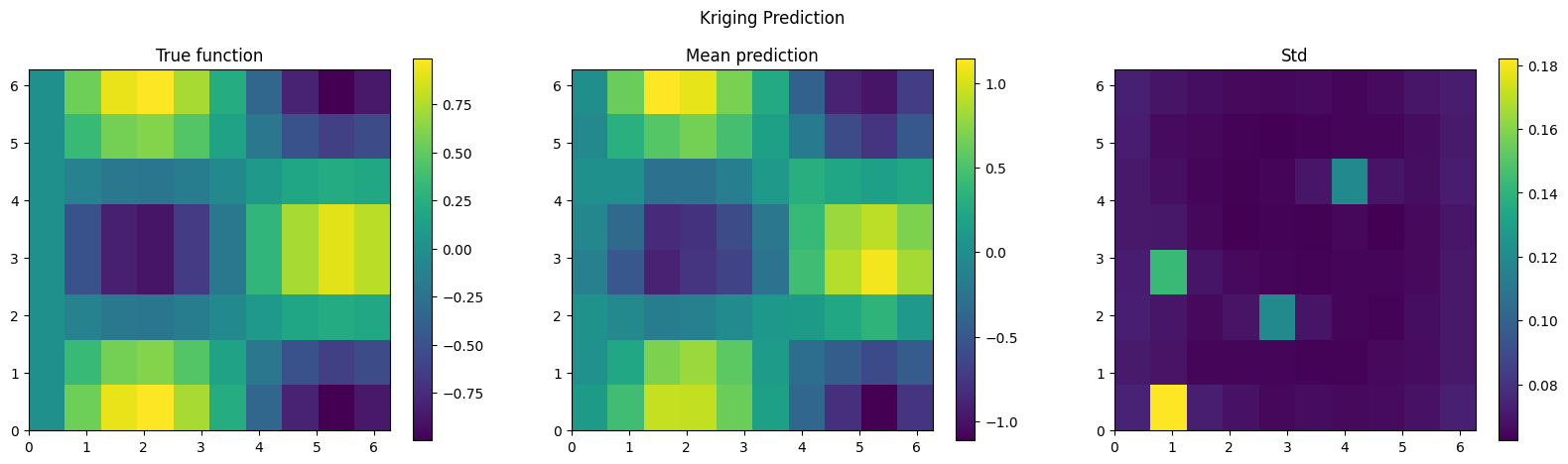
# ATTENTION: this cell might take a while to run
# fit the model
nuts_kernel = NUTS(model)
mcmc = MCMC(nuts_kernel, num_samples=10000, num_warmup=2000, num_chains=2, chain_method='parallel', progress_bar=False)
mcmc.run(jax.random.PRNGKey(42), jnp.array(x_2d), jnp.array(obs_idx), jnp.array(y_obs), kernel_func=rbf_kernel, lengthcsale=1.0)
# exatract posterior
posterior_samples = mcmc.get_samples()
f_posterior = posterior_samples['f']
f_mean = jnp.mean(f_posterior, axis=0)
f_std = jnp.std(f_posterior, axis=0)
f_mean_2d = f_mean.reshape(xx.shape)
f_std_2d = f_std.reshape(xx.shape)
plt.figure(figsize=(20, 5))
plt.subplot(1, 3, 1)
plt.imshow(f_true, extent=(0, 2*np.pi, 0, 2*np.pi), origin='lower', cmap='viridis')
plt.colorbar()
plt.title('True function')
plt.subplot(1, 3, 2)
plt.imshow(f_mean_2d, extent=(0, 2*np.pi, 0, 2*np.pi), origin='lower', cmap='viridis')
plt.colorbar()
plt.title('Mean prediction')
plt.subplot(1, 3, 3)
plt.imshow(f_std_2d, extent=(0, 2*np.pi, 0, 2*np.pi), origin='lower', cmap='viridis')
plt.colorbar()
plt.title('Std')
plt.suptitle('Kriging Prediction')
plt.show()
xmin = np.min([np.min(f_true_flat), np.min(f_mean)])
xmax = np.max([np.max(f_true_flat), np.max(f_mean)])
ymin = xmin
ymax = xmax
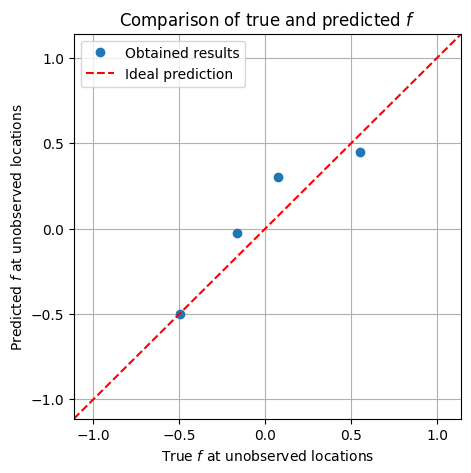
plt.figure(figsize=(5, 5))
plt.plot(f_true_flat[skip_idx_1d], f_mean[skip_idx_1d], 'o', label='Obtained results')
plt.plot([xmin, xmax], [ymin, ymax], color='red', linestyle='--', label='Ideal prediction')
plt.xlabel('True $f$ at unobserved locations')
plt.ylabel('Predicted $f$ at unobserved locations')
plt.xlim(xmin, xmax)
plt.ylim(ymin, ymax)
plt.title('Comparison of true and predicted $f$')
plt.legend()
plt.grid(True)
plt.show()
Note that in the examples above we used the Gaussian likelihood for simplicity. But in applied research this wil seldomly be the case.

