Gaussian Processes: priors#
Nonparametric models#
So far in this course, all the models we have built were regressions, working in the supervised learning setting and using parametric models. We tried to describe functions with unknown parameters using Bayesian formalism.
Today we will introduce nonparametric models. The term nonparametric means that the shape of the functions is determined by the data as opposed to parametric functions which are defined by a typically fixed set of parameters. In nonparametric models the number of parameters grows with the number of data points. Nonparametric functions are extremely flexible since their shape adapts to the underlying patterns in the data.
Nonparametric models are commonly based on kernel functions. A kernel function \(k(x, x′)\) is a map of a pair of inputs \(x\) and \(x′ \in \mathcal{X}\) with input domain \(\mathcal{X}\) into real numbers
characterizing the similarity of the pair of inputs \(x, x'\).
Gaussian processes - preview#
Gaussian processes (GPs) [Williams and Rasmussen, 2006] are an example of nonparametric models. They are powerful and flexible statistical models commonly used in GLMMs as random effects where they act as latent variable.
A GP can be defined as a collection of random variables, any finite subset of which has a joint multivariate normal distribution typically represented as
where \(\mu(x) \) is the mean function and \( k(x, x') \) is the covariance function.
A GP defines a distribution over functions rather than individual points.
GPs can model a wide variety of functions without assuming a specific functional form. This makes them particularly useful when dealing with complex or unknown relationships in data.
GPs are inherently Bayesian models, meaning they provide a principled way to quantify uncertainty in predictions. This is achieved by representing the posterior distribution over functions given the observed data.
The choice of kernel function determines the behavior and characteristics of the GP. Common kernel functions include the radial basis function (RBF), also known as the squared exponential kernel, and the Matérn kernel, among others. These kernels encode assumptions about the smoothness and structure of the underlying function. We will see specific examples of kernels in this lecture.
While GPs offer many advantages, they can be computationally intensive, especially as the size of the dataset grows. Various approximation methods such as sparse GPs and approximate inference techniques are used to scale Gaussian processes to larger datasets.
This was a preview. We will return to the formal definition once again later in this lecture.
Now let’s build the ingredients which we need to understand Gaussian processes step by step.
From Univariate to Multivariate Gaussians#
Univariate Normal distribution#
Recall from previous chapters, the univariate normal distribution has PDF
As before, the notation here will be \(y \sim \mathcal{N}(\mu, \sigma^2)\),
Reparametrization#
Note, that in order to sample variable \(y\) we could first sample a standard Normal variable \(z \sim \mathcal{N}(0,1)\), and then perform a transformation of this variable
Group Task
Prove that \(y:= \mu + \sigma z\) is indeed distributed as \(\mathcal{N}(\mu, \sigma^2).\)
Hint: What you need to show for that is that \(\mathbb{E}[y] = \mu\) and \(\text{Var}[y] = \sigma^2\). To conclude, make an argument about why \(y\) is normally distributed.
Multivariate Normal distribution#
In the multivariate case, instead of using scalar mean and variance parameters \(\mu, \sigma^2 \in \mathbb{R}\), we need to specify a vector mean \(\mu \in \mathbb{R}^d\) and covariance matrix \(K \in \mathbb{R}^{d \times d}.\) To write that variable \(y\) follows a multivariate mormal distribution, we use notation
Bivariate case#
‘Bivariate’ means \(d=2\). Hence,
Here \(\mu_i\) is the mean of component \(y_i\), \(\sigma_i^2\) is the variance for the \(i\)-th dimension, and \(\rho_{ij} = \rho_{ji}\) is the correlation between the \(i\)-th and \(j\)-th dimensions:
The covariance matrix tells us how the “ball” of random variables is stretched and rotated in space. Let’s visualise a few examples.
import numpy as np
#from scipy.stats import norm
import jax.numpy as jnp
from jax import jit
import matplotlib.pyplot as plt
from matplotlib.colors import Normalize
from matplotlib.cm import ScalarMappable
import matplotlib.gridspec as gridspec
import numpyro.distributions as dist
/opt/hostedtoolcache/Python/3.11.11/x64/lib/python3.11/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
def plot_2d_gp(mu1, mu2, rho=0, sigma1=1, sigma2=1):
# generate data points from the 2D Gaussian distribution
mu = np.array([mu1, mu2])
covariance = jnp.array([[sigma1**2, rho*sigma1*sigma2],[rho*sigma1*sigma2, sigma2**2]])
num_samples = 1000
data = np.random.multivariate_normal(mu, covariance, num_samples)
# calculate marginal distributions
y_values = np.linspace(-3, 3, 1000)
normal1 = dist.Normal(loc= mu[0], scale =np.sqrt(covariance[0, 0]))
normal2 = dist.Normal(loc= mu[1], scale =np.sqrt(covariance[1, 1]))
marginal_y1 = jnp.exp(normal1.log_prob(y_values))
marginal_y2 = jnp.exp(normal1.log_prob(y_values))
# create figure
fig = plt.figure(figsize=(8, 6))
gs = fig.add_gridspec(3, 3)
# main plot (2D Gaussian distribution)
ax_main = fig.add_subplot(gs[1:3, :2])
ax_main.scatter(data[:, 0], data[:, 1], alpha=0.5, label='2d Gaussian')
ax_main.set_xlabel('$y_1$')
ax_main.set_ylabel('$y_2$')
ax_main.legend()
ax_main.grid(True)
# marginal X plot
ax_marginal_x = fig.add_subplot(gs[0, :2], sharex=ax_main)
ax_marginal_x.plot(y_values, marginal_y1, label='Marginal $y_1$', color='orangered')
ax_marginal_x.legend()
ax_marginal_x.grid(True)
# marginal Y plot
ax_marginal_y = fig.add_subplot(gs[1:3, 2], sharey=ax_main)
ax_marginal_y.plot(marginal_y2, y_values, label='Marginal $y_2$', color='purple')
ax_marginal_y.legend()
ax_marginal_y.grid(True)
plt.tight_layout()
plt.show()
# parameters for the 2D Gaussian distribution
mu1 = 0
mu2 = 0
rho = 0.1
plot_2d_gp(mu1, mu2, rho)
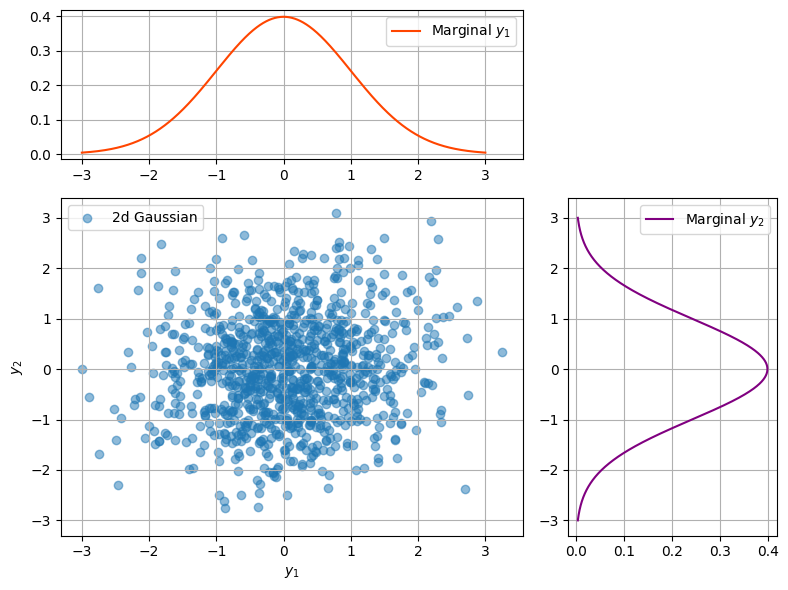
Group Task
Loop over several values of \(\rho\). How does the distribution change?
\(d>2\)#
Visualising the distribution in dimensions higher than 2 is harder.
Still, in 2d we can use indices of components of \(y\): 1 for \(y_1\) and 2 for \(y_2\) to make the plots.
# parameters for the 2D Gaussian distribution
mu = np.array([0, 0]) # mean
covariance = np.array([[1, 0.5], [0.5, 1]]) # covariance matrix
num_samples = 10
data = np.random.multivariate_normal(mu, covariance, num_samples)
# extract values at indices 1 and 2 for each sample
index_1 = 1
index_2 = 2
values_1 = data[:, index_1 - 1]
values_2 = data[:, index_2 - 1]
# generate a color map
cmap = plt.get_cmap('viridis')
norm = Normalize(vmin=0, vmax=num_samples - 1)
scalar_map = ScalarMappable(norm=norm, cmap=cmap)
plt.figure(figsize=(4, 6))
for i in range(num_samples):
color = scalar_map.to_rgba(i)
plt.plot([1, 2], [values_1[i], values_2[i]], color=color, marker='o', linestyle='-', label=f'sample {i}')
plt.xticks([1, 2], ['1', '2']) # Set x-axis ticks to only 1 and 2
plt.xlabel('index')
plt.ylabel('value')
plt.title(f'Values at indices 1 and 2 \nfor each sample ({num_samples})')
plt.tight_layout()
plt.legend(loc='upper right', bbox_to_anchor=(1.8, 1))
plt.grid(True)
plt.show()
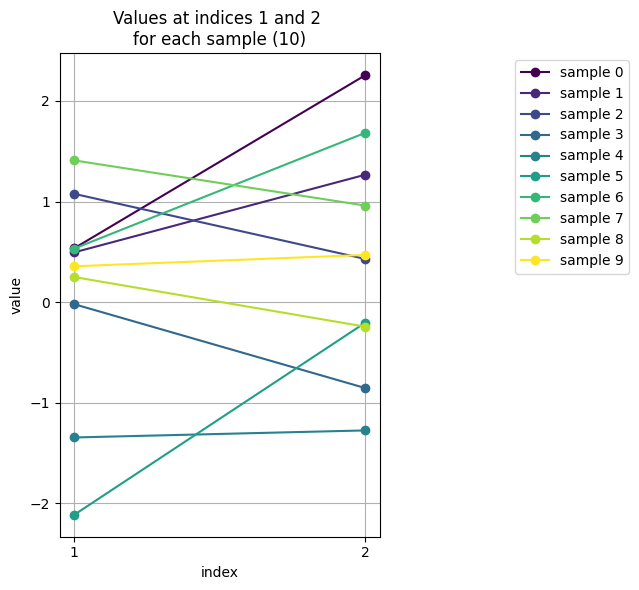
Can we generalise this approach to visualise data of higher dimensionality \(d\)?
def plot_values(data, d):
num_samples = data.shape[0]
indices = list(range(1, d + 1)) # indices to plot
values = [data[:, idx - 1] for idx in indices]
# color map
cmap = plt.get_cmap('viridis')
norm = Normalize(vmin=0, vmax=num_samples - 1)
scalar_map = ScalarMappable(norm=norm, cmap=cmap)
plt.figure(figsize=(6, 6))
for i in range(num_samples):
color = scalar_map.to_rgba(i)
plt.plot(indices, [values[j][i] for j in range(len(indices))], color=color, marker='o', linestyle='-', label=f'sample {i}')
plt.xticks(indices, [str(idx) for idx in indices])
plt.xlabel('index')
plt.ylabel('value')
plt.title(f'Values at indices 1 to {d} for eachsSample ({num_samples})')
plt.tight_layout()
plt.legend(loc='upper right', bbox_to_anchor=(1.3, 1))
plt.grid(True)
plt.show()
# parameters for the multivariate Gaussian distribution
# mean vector
mu = np.zeros(5)
# covariance matrix
covariance = np.array([[1, 0.8, 0.7, 0.4, 0.1],
[0.8, 1, 0.5, 0.3, 0.2],
[0.7, 0.5, 1, 0.2, 0.1],
[0.4, 0.3, 0.2, 1, 0.1],
[0.1, 0.2, 0.1, 0.1, 1]])
# generate data from multivariate normal distribution
# number of components in the vector
d = 5
# number fo samples
num_samples = 10
data = np.random.multivariate_normal(mu, covariance, num_samples)
plot_values(data, d)
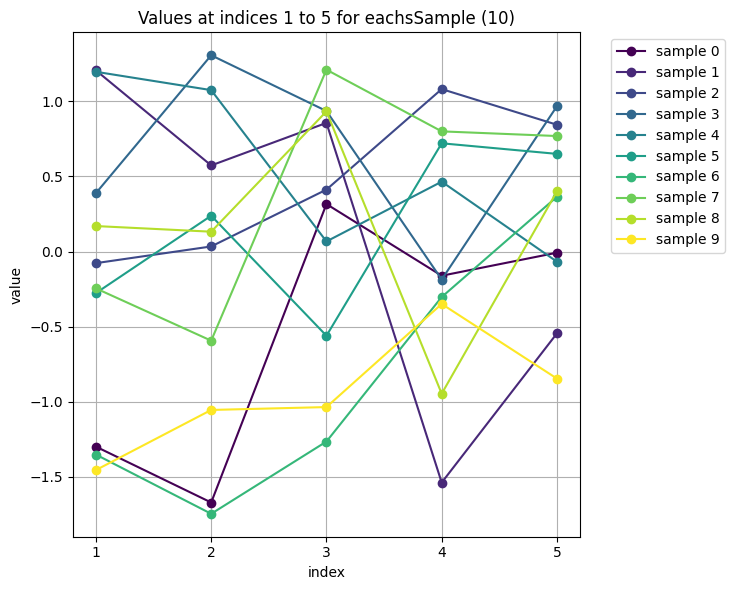
In the example above we coded up the covariance matris by hand. It is time to look into what covariance matrices might be more meaningful.
Gaussian process - revisiting the definition#
Let’s revisit what a Gaussian process is.
A stochastic process is a collection of random variables \(f=f(x)\) indexed by some variable \(x \in \mathcal{X}\).
A Gaussian process is a stochastic process \(f: \mathcal{X} \to \mathbb{R}\), such that its every finite realisation \( [f(x_1), ..., f(x_d)]^T\) follows a multivariate normal dsitribution
Hence, a Gaussian process is fully specified by its mean and covariance functions \(\mu(x), k(x,x'):\)
where
and
Typically, for the mean function we chose one of the following options \(\mu(x)=0, \mu(x)=c, \mu(x)=\beta^Tx\).
Kernels#
Covariance function must be a positive semi-definite function in order to lead to valid covariance matrices.
A kernel \(k: \mathcal{X} \times \mathcal{X} \to \mathbb{R}\) is positive semi-definite, if for any finite collection \(x= (x_1, ..., x_d)\) the matrix \(k_{xx}\) with \([k_{xx}]_{ij}=k(x_i, x_j)\) is positive semi-definite.
A symmetric matrix \(A \in \mathbb{R}^{N \times N}\) is called positive semi-definite if
for any \(v \in \mathbb{R}^d.\)
Kernel functions \(k(x, x′)\) encode our prior beliefs about data-generating latent functions. These typically include continuity, smoothness (differentiability),periodicity, stationarity, and so on.
Covariance functions typically have hyperparameters that we aim to learn from data.
Let us explore some typical covariance functions.
Radial basis function (RBF)#
The squared exponential kernel, also known as the Gaussian kernel or the radial basis function (RBF) kernel is one of the most commonly used kernels in Gaussian processes due to its smoothness. It is defined as
where \(\sigma^2\) is the variance parameter (also called the amplitude), \(l\) is the lengthscale parameter and \(\|x_i - x_j\|\) is the Euclidean distance between the points \(x_i\) and \(x_j\).
This kernel assigns high similarity (and hence high covariance) to points that are close to each other in the input space and low similarity (and low covariance) to points that are far apart. The parameters \(\sigma^2\) and \(l\) control the overall variance and the rate at which the covariance decreases with distance, respectively.
def rbf_kernel(x1, x2, sigma=1.0, lengthscale=1.0, jitter=1e-6):
"""
compute the Radial Basis Function (RBF) kernel matrix between two sets of points.
args:
- x1 (array): array of shape (n1, d) representing the first set of points
- x2 (array): array of shape (n2, d) representing the second set of points
- sigma (float): variance parameter
- length_scale (float): length-scale parameter
- jitter (float): amall positive value added to the diagonal elements
returns:
- K (array): kernel matrix of shape (n1, n2)
"""
sq_dist = jnp.sum(x1**2, axis=1).reshape(-1, 1) + jnp.sum(x2**2, axis=1) - 2 * jnp.dot(x1, x2.T)
K = sigma**2 * jnp.exp(-0.5 / lengthscale**2 * sq_dist)
# add jitter to ensure positive definiteness. This is a purely numeric trick
K += jitter * jnp.eye(K.shape[0])
return K
Let us visualise several samples of the GP with RBF kernel with a fixed lengthscale:
# define parameters
n_points = 50
num_samples = 10
sigma = 1.0
lengthscale = 1.0
jitter = 1e-4
# generate random input data
x = jnp.linspace(0, 10, n_points).reshape(-1, 1)
# compute covariance matrix using RBF kernel function
K = rbf_kernel(x, x, sigma=sigma, lengthscale=lengthscale, jitter=jitter)
# color map
cmap = plt.get_cmap('viridis')
norm = Normalize(vmin=0, vmax=num_samples - 1)
scalar_map = ScalarMappable(norm=norm, cmap=cmap)
mu = np.zeros(n_points) # mean vector
data = np.random.multivariate_normal(mu, K, num_samples)
plt.figure(figsize=(8, 6))
for i in range(num_samples):
color = scalar_map.to_rgba(i)
plt.plot(x, data[i,:], linestyle='-', color=color, label=f'sample {i}')
plt.xlabel('$x$')
plt.ylabel('$f(x)$')
plt.title(f'RBF kernel with lengthscale={lengthscale}')
plt.tight_layout()
plt.legend(loc='upper right', bbox_to_anchor=(1.3, 1))
plt.grid(True)
plt.show()
/tmp/ipykernel_2850/1332337364.py:20: RuntimeWarning: covariance is not symmetric positive-semidefinite.
data = np.random.multivariate_normal(mu, K, num_samples)
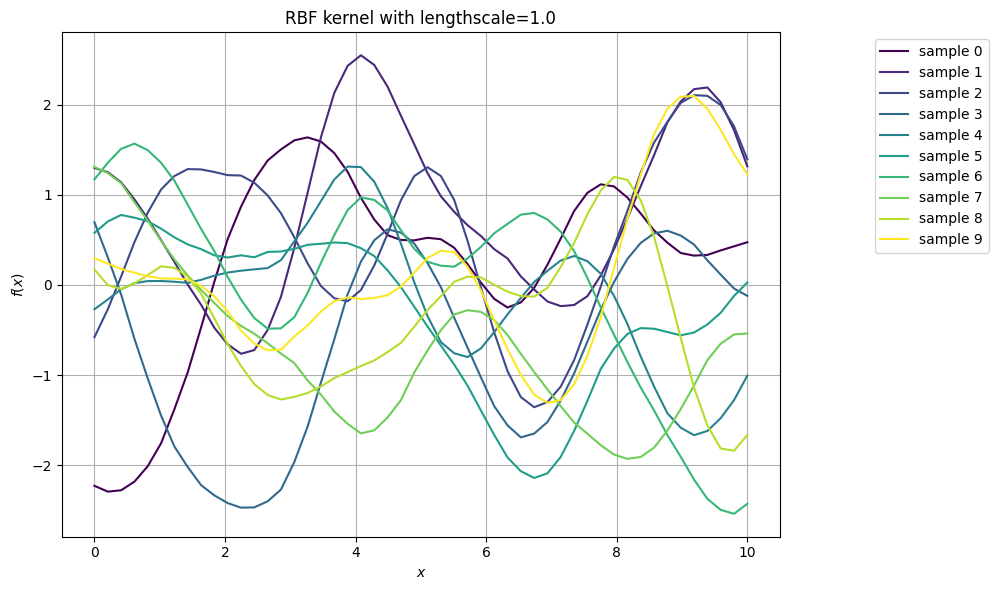
How do curves with different lengthscales look like?
# lengthscale values for each sample
lengthscales = np.array([0.1, 0.3, 0.5, 0.7, 1.0, 1.5, 5.0])
plt.figure(figsize=(8, 6))
for i, lengthscale in enumerate(lengthscales):
# compute covariance matrix using RBF kernel function with different lengthscale for each sample
K = rbf_kernel(x, x, sigma=sigma, lengthscale=lengthscale, jitter=jitter)
# draw samples from multivariate Gaussian distribution
mu = np.zeros(n_points)
data = np.random.multivariate_normal(mu, K)
# generate a color corresponding to the lengthscale
color = cmap(i / (len(lengthscales) - 1))
# plot sample
plt.plot(x, data, linestyle='-', color=color, label=f'lengthscale={lengthscale:.2f}')
plt.xlabel('$x$')
plt.ylabel('$f(x)$')
plt.title(f'RBF kernel with varying lengthscales')
plt.tight_layout()
plt.legend(loc='upper right', bbox_to_anchor=(1.3, 1))
plt.grid(True)
plt.show()
/tmp/ipykernel_2850/2293107768.py:14: RuntimeWarning: covariance is not symmetric positive-semidefinite.
data = np.random.multivariate_normal(mu, K)
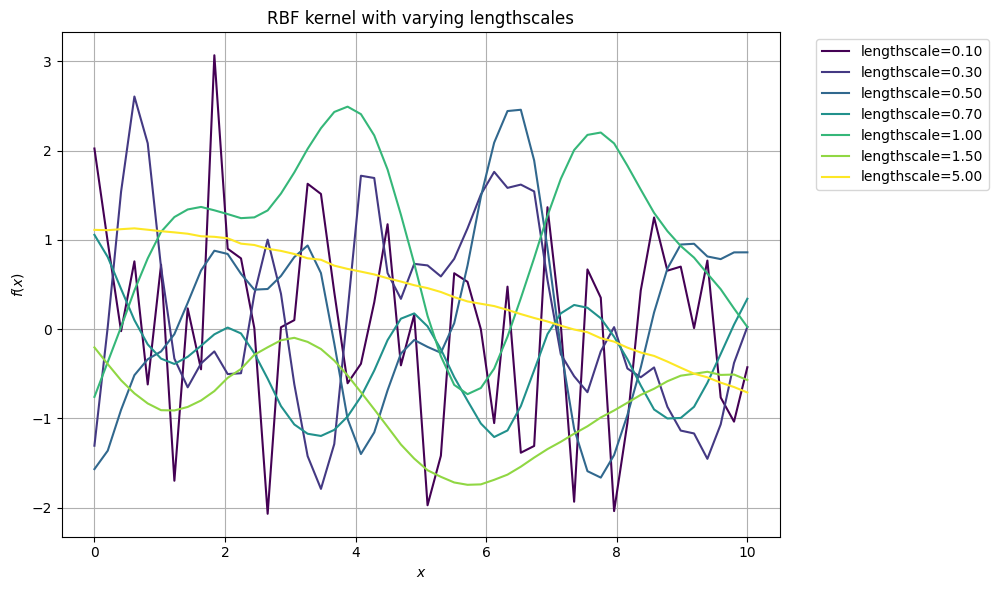
Group Task
How can you describe this dependence of the trajectories on the lengthscale?
Group Task
Explore this interactive visualisation.
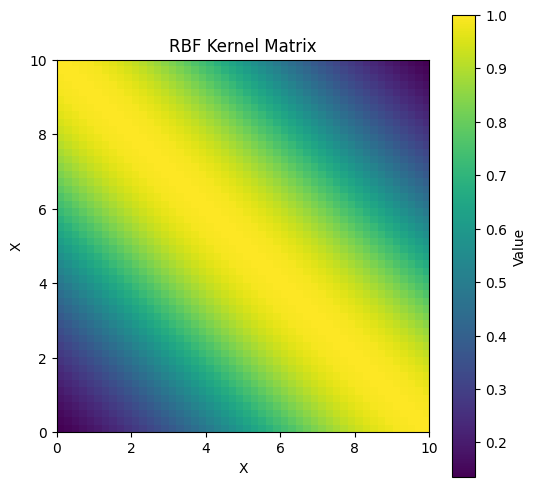
Apart from visualising trajectories, we can also plot the covariance matrix:
# visualise covariance matrix
plt.figure(figsize=(6, 6))
plt.imshow(K, cmap='viridis', extent=[0, 10, 0, 10])
plt.colorbar(label='Value')
plt.title('RBF Kernel Matrix')
plt.xlabel('X')
plt.ylabel('X')
plt.show()
We will need to plot many more GP trajectories. Let’s make a wrapper:
def plot_gp_samples(x, K, ttl="", num_samples=5):
num_points = len(x)
samples = np.random.multivariate_normal(mean=np.zeros(num_points), cov=K, size=num_samples)
# plot the samples
plt.figure(figsize=(8, 6))
for i in range(num_samples):
color = cmap(i / (len(lengthscales) - 1))
plt.plot(x, samples[i], label=f'Sample {i}', color=color)
plt.xlabel('x')
plt.ylabel('f(x)')
plt.title(ttl)
plt.legend()
plt.tight_layout()
plt.legend(loc='upper right', bbox_to_anchor=(1.3, 1))
plt.grid(True)
plt.show()
## test the function
#n_points = 50
#sigma = 1.0
#lengthscale = 1.0
#jitter = 1e-4
## generate random input data
#x = jnp.linspace(0, 10, n_points).reshape(-1, 1)
#K = rbf_kernel(x, x, sigma=sigma, lengthscale=lengthscale, jitter=jitter)
#plot_gp_samples(x, K, ttl="RBF kernel demonstration")
Matérn kernels#
The Matérn kernel is another popular choice in Gaussian processes. It is a flexible covariance function that is able to capture different levels of smoothness in the data. The Matérn kernel is defined as:
where \(k(x_i, x_j)\) represents the covariance between two data points \(x_i\), \(x_j\), \(\sigma^2\) is the variance parameter, \(\nu\) is the smoothness parameter, typically a positive half-integer (\(\nu = 1/2, 3/2, 5/2\)), \(l\) is the length-scale parameter, \(\|x_i - x_j\|\) is the Euclidean distance between the points \(x_i\) and \(x_j\), \(K_\nu\) is the modified Bessel function of the second kind of order \(\nu\), \(\Gamma\) is the gamma function.
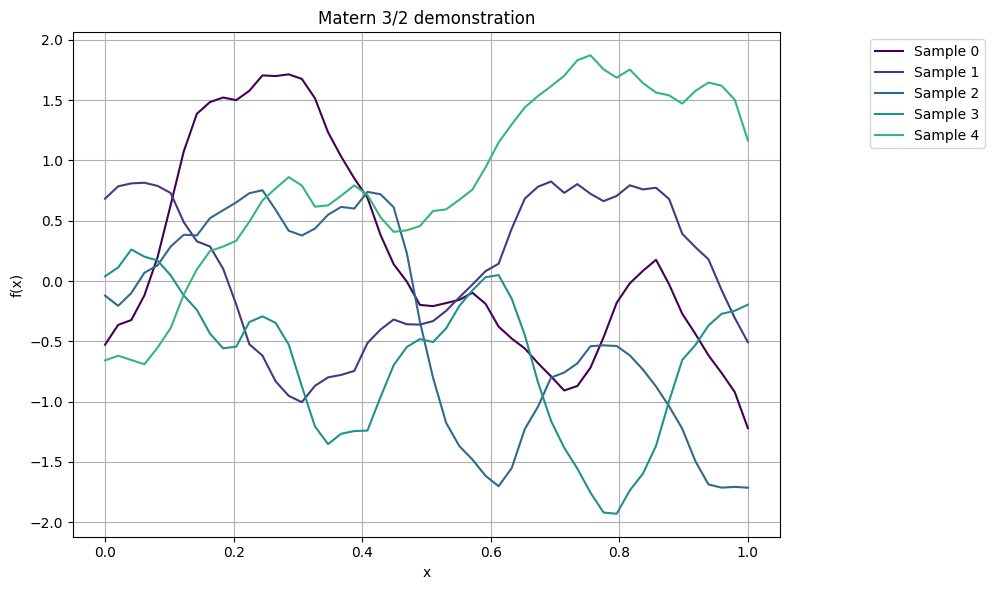
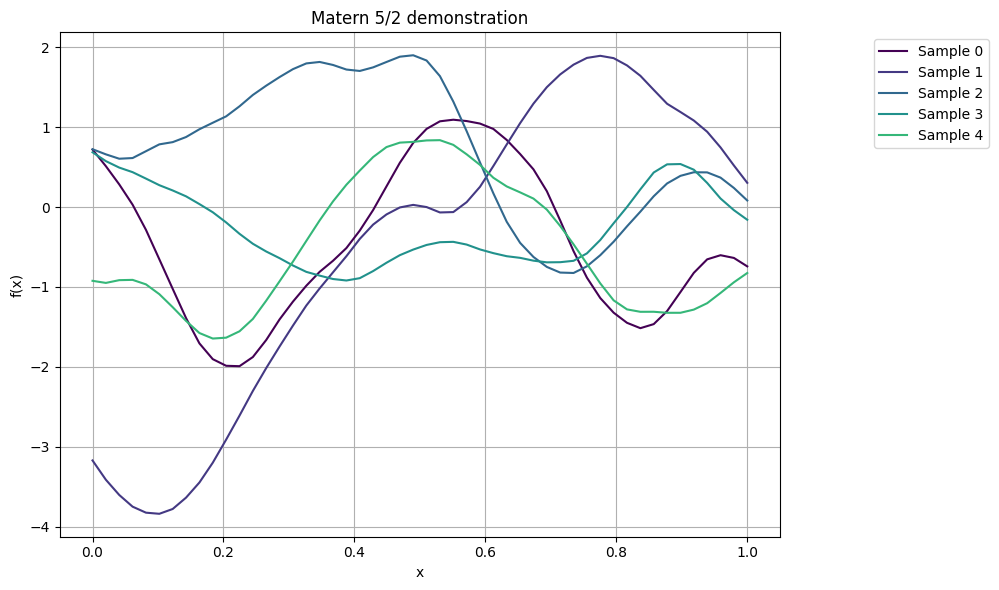
Matérn 3/2 and 5/2#
The Matérn kernel with \(\nu=3/2\) is a particular case of the Matérn family of kernels and is defined as
The Matérn kernel with \(\nu=5/2\) is another member of the Matérn family of kernels. The formula for the Matérn-5/2 kernel is given by
def matern32_kernel(x1, x2, sigma=1.0, lengthscale=1.0):
"""
compute the Matérn-3/2 kernel matrix between two sets of points.
args:
- x1 (array): array of shape (n1, d) representing the first set of points
- x2 (array): array of shape (n2, d) representing the second set of points
- sigma (float): variance
- length_scale (float): length-scale
returns:
- K (array): kernel matrix of shape (n1, n2)
"""
dist = jnp.sqrt(jnp.sum((x1[:, None] - x2) ** 2, axis=-1))
arg = dist / lengthscale
return sigma**2 * (1 + jnp.sqrt(3) * arg) * jnp.exp(- jnp.sqrt(3) * arg)
# compile the kernel function for better performance
matern32_kernel = jit(matern32_kernel)
def matern52_kernel(x1, x2, sigma=1.0, lengthscale=1.0):
"""
compute the Matérn-5/2 kernel matrix between two sets of points
args:
- x1 (array): array of shape (n1, d) representing the first set of points
- x2 (array): array of shape (n2, d) representing the second set of points
- sigma (float): variance
- length_scale (float): length-scale
returns:
- K (array): kernel matrix of shape (n1, n2)
"""
dist = jnp.sqrt(jnp.sum((x1[:, None] - x2) ** 2, axis=-1))
arg = dist / lengthscale
return sigma**2 * (1 + jnp.sqrt(5) * arg + 5/3 * arg**2) * jnp.exp(-jnp.sqrt(5) * arg)
# compile the kernel function for better performance
matern52_kernel = jit(matern52_kernel)
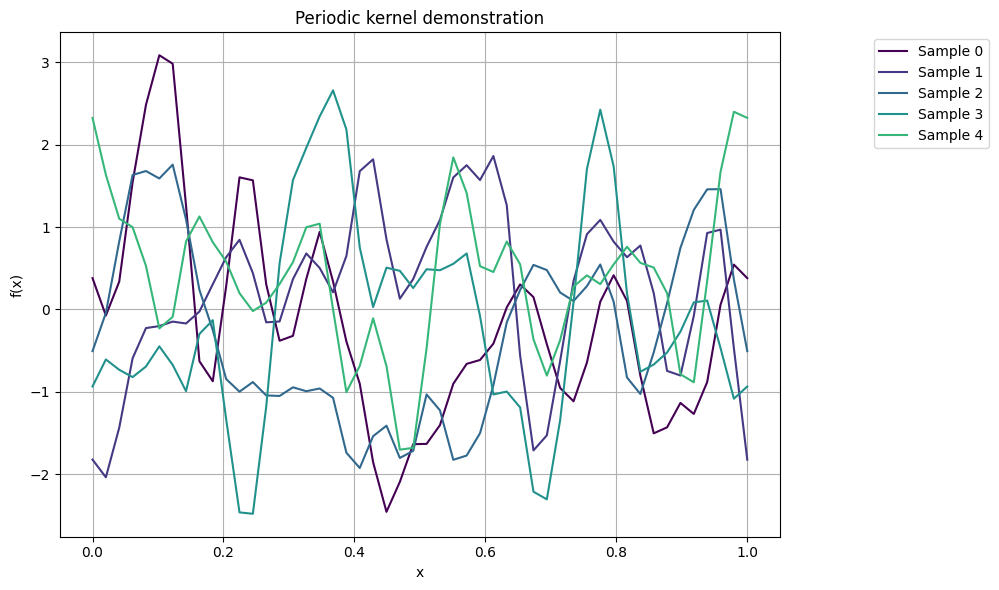
Periodic kernel#
The periodic kernel is commonly used in Gaussian processes to model periodic patterns in data. The formula for the periodic kernel is given as
def periodic_kernel(x1, x2, sigma=1.0, lengthscale=1.0, period=1.0):
"""
compute the periodic kernel matrix between two sets of points.
args:
- x1 (array): array of shape (n1, d) representing the first set of points.
- x2 (array): array of shape (n2, d) representing the second set of points.
- sigma (float): variance
- length_scale (float): length-scale parameter
- period (float): periodicity parameter
returns:
- K (array): kernel matrix of shape (n1, n2)
"""
dist = jnp.sqrt(jnp.sum((x1[:, None] - x2) ** 2, axis=-1))
return sigma**2 * jnp.exp(-2 * jnp.sin(jnp.pi * dist/ period)**2 / lengthscale**2)
# compile the kernel function for better performance
periodic_kernel = jit(periodic_kernel)
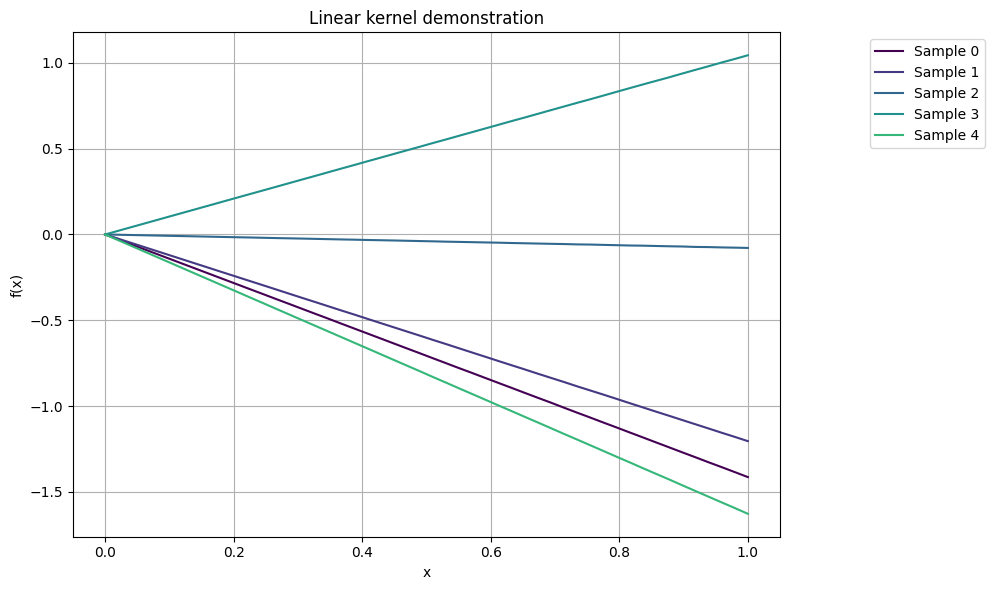
Linear kernel#
The linear kernel, also known as the dot product kernel, is one of the simplest kernels. It computes the covariance between two data points as the inner product of their feature vectors. The formula for the linear kernel is given as
def linear_kernel(x1, x2, sigma =1.0):
"""
Compute the linear kernel matrix between two sets of points.
args:
- x1 (array): array of shape (n1, d) representing the first set of points
- x2 (array): array of shape (n2, d) representing the second set of points
returns:
- K (array): kernel matrix of shape (n1, n2)
"""
return sigma**2 * jnp.dot(x1, x2.T)
# compile the kernel function for better performance
linear_kernel = jit(linear_kernel)
Now let’s sisualise these kernels.
# test the function
n_points = 50
sigma = 1.0
lengthscale = 0.2
jitter = 1e-4
# input locations
x = jnp.linspace(0, 1, n_points).reshape(-1, 1)
# compute covariance matrix using RBF kernel function
K32 = matern32_kernel(x, x, sigma=sigma, lengthscale=lengthscale)
K52 = matern52_kernel(x, x, sigma=sigma, lengthscale=lengthscale)
K_per = periodic_kernel(x, x, sigma=sigma, lengthscale=lengthscale)
K_lin = linear_kernel(x, x, sigma=sigma)
plot_gp_samples(x, K32, ttl="Matern 3/2 demonstration")
plot_gp_samples(x, K52, ttl="Matern 5/2 demonstration")
plot_gp_samples(x, K_per, ttl="Periodic kernel demonstration")
plot_gp_samples(x, K_lin, ttl="Linear kernel demonstration")
/tmp/ipykernel_2850/2518918070.py:4: RuntimeWarning: covariance is not symmetric positive-semidefinite.
samples = np.random.multivariate_normal(mean=np.zeros(num_points), cov=K, size=num_samples)
Task 26
Implement Matern-1/2 kernel:
plot function draws,
plot covariance matrix
Implement the rational quadratic kernel:
plot function draws,
plot covariance matrix
Making new kernels#
Given two kernels \(k_1(x, x')\) and \(k_2(x, x')\), we can create a valid new kernel using any of the following methods [Murphy, 2023]:
\(k(x, x') = c k_1(x, x'), \quad c>0\)
\(k(x, x') = f(x) k_1(x, x') f(x')\) for any function \(f\)
\(k(x, x') = \exp(k_1(x, x'))\)
\(k(x, x') = x^\top A x'\) for any \(A \ge 0\)
\(k(x, x') = k_1(x, x') + k_2(x, x')\)
\(k(x, x') = k_1(x, x') k_2(x, x')\)
I.e. kernels can be combined to make new kernels.
Cholesky decomposition and reparametrization#
Recall the reparametrization trick for \(d=1\). Can we perform a similar computation in \(d>1\)?
Cholesky decomposition is a numerical method used to decompose a positive definite matrix into a lower triangular matrix and its conjugate transpose: for a positive definite matrix \(A\) it has the form
where \(L\) is a lower triangular matrix, \(L^T\) is the transpose of \(L\).
Cholesky decomposition is particularly useful because it provides a computationally efficient way to solve linear systems of equations, including inverting matrices and calculating determinants.
In the Gaussian processes context, Cholesky decomposition is commonly used to generate samples from a multivariate Gaussian distribution. Here we find the Cholesky decomposition of the covariance matrix \(K\) which yields a lower triangular matrix \(L\):
By multiplying this lower triangular matrix with a vector of independent standard normals \(z\), we can generate samples from the Gaussian process while ensuring that the resulting samples have the desired covariance structure encoded by the covariance matrix \(K\). Hence, we can either sample directly
or use the reparametrization
Task 27
Show that \(f:= \mu + Lz\) is indeed distributed as \(\mathcal{N}(\mu, K)\).

